Rename#
Rename and reorient the chromosome, according to a monoploid or closely relative species's genome.
Rename phased assembly from phasing mode#
The phasing mode of C-Phasing has been clustering the chromsomes into the same homologous and oriented them to parallel.
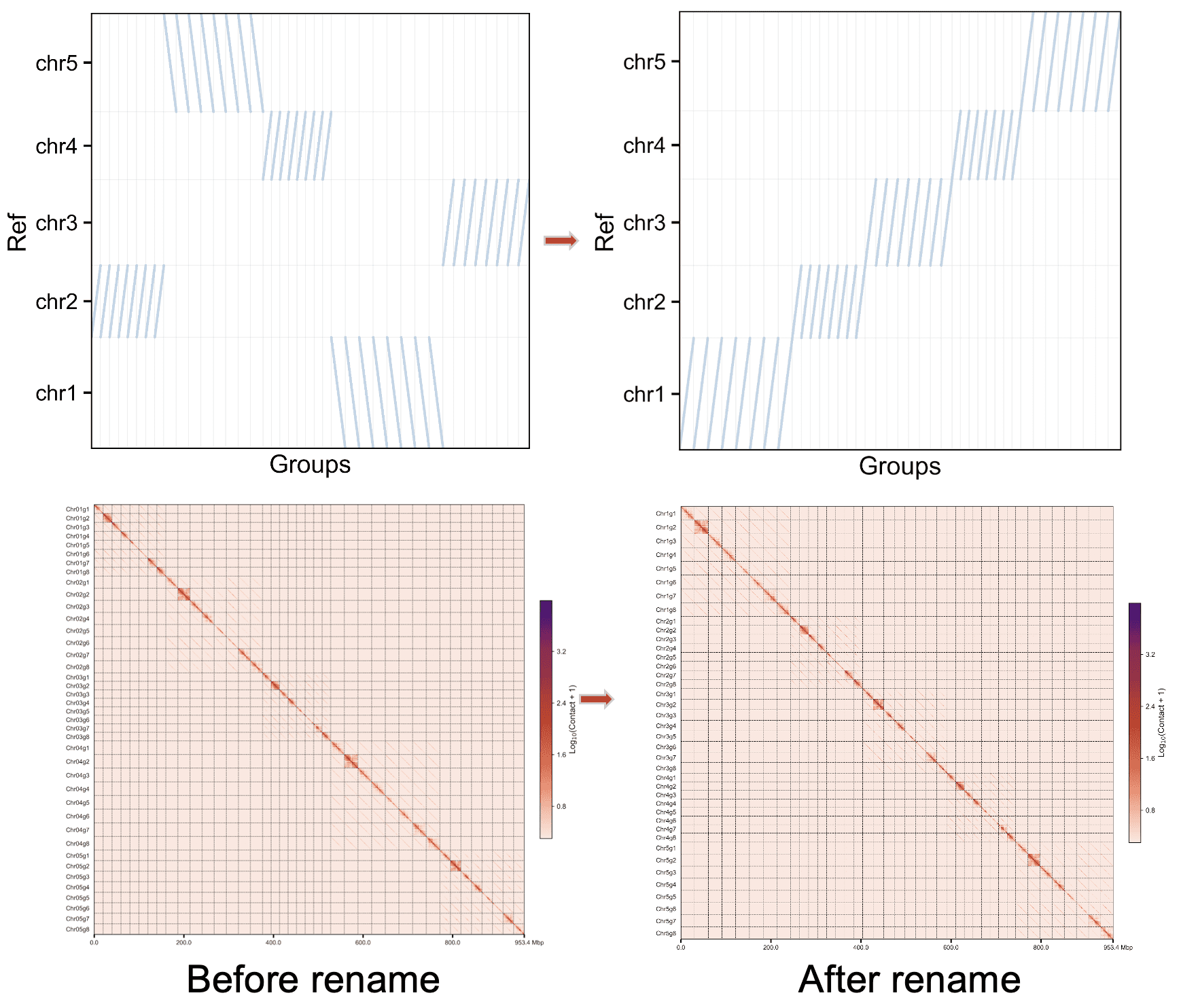
Rename all of the chromosomes#
Rename assembly from basal or basal_withprune mode, with the --unphased parameter.
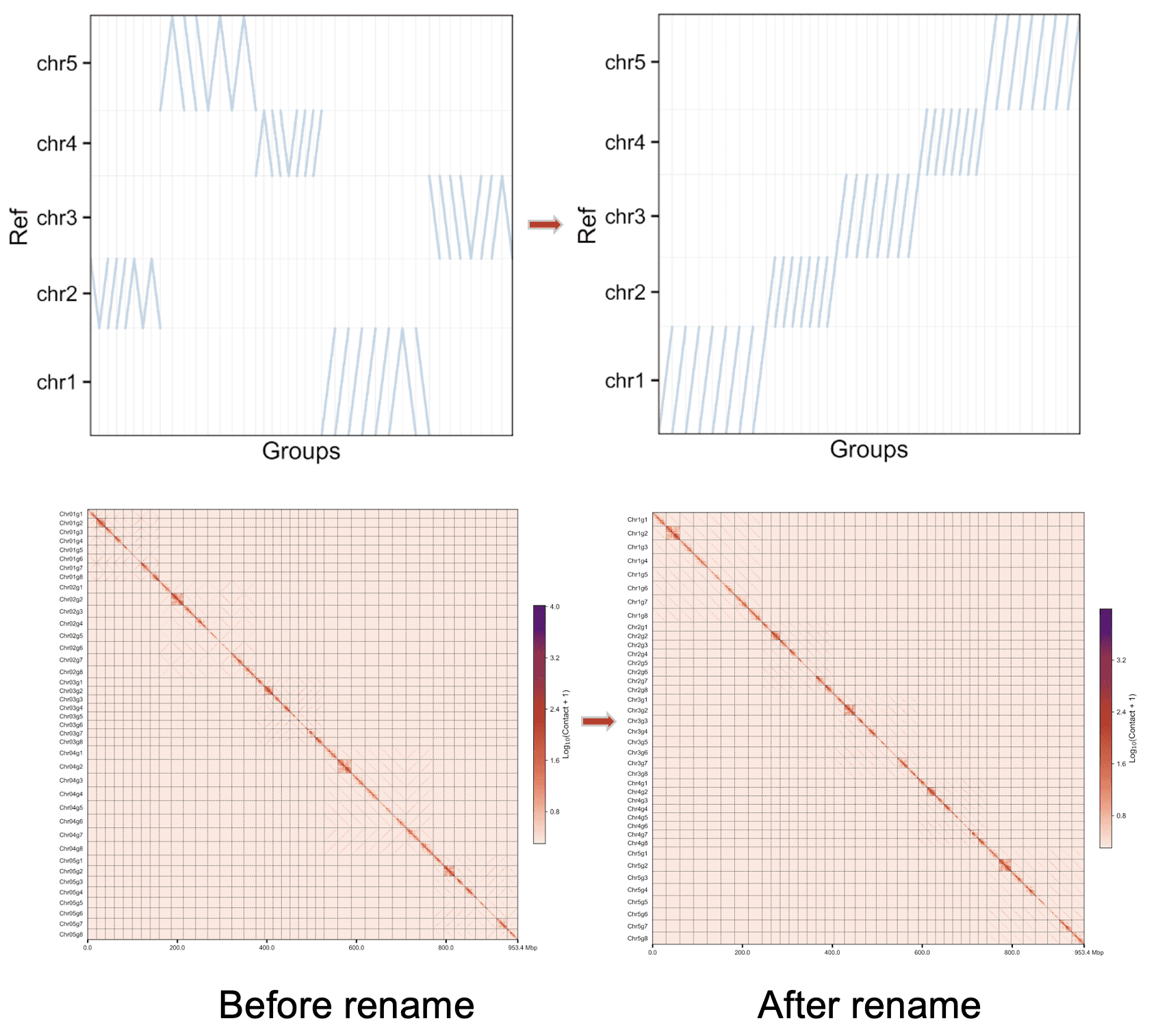
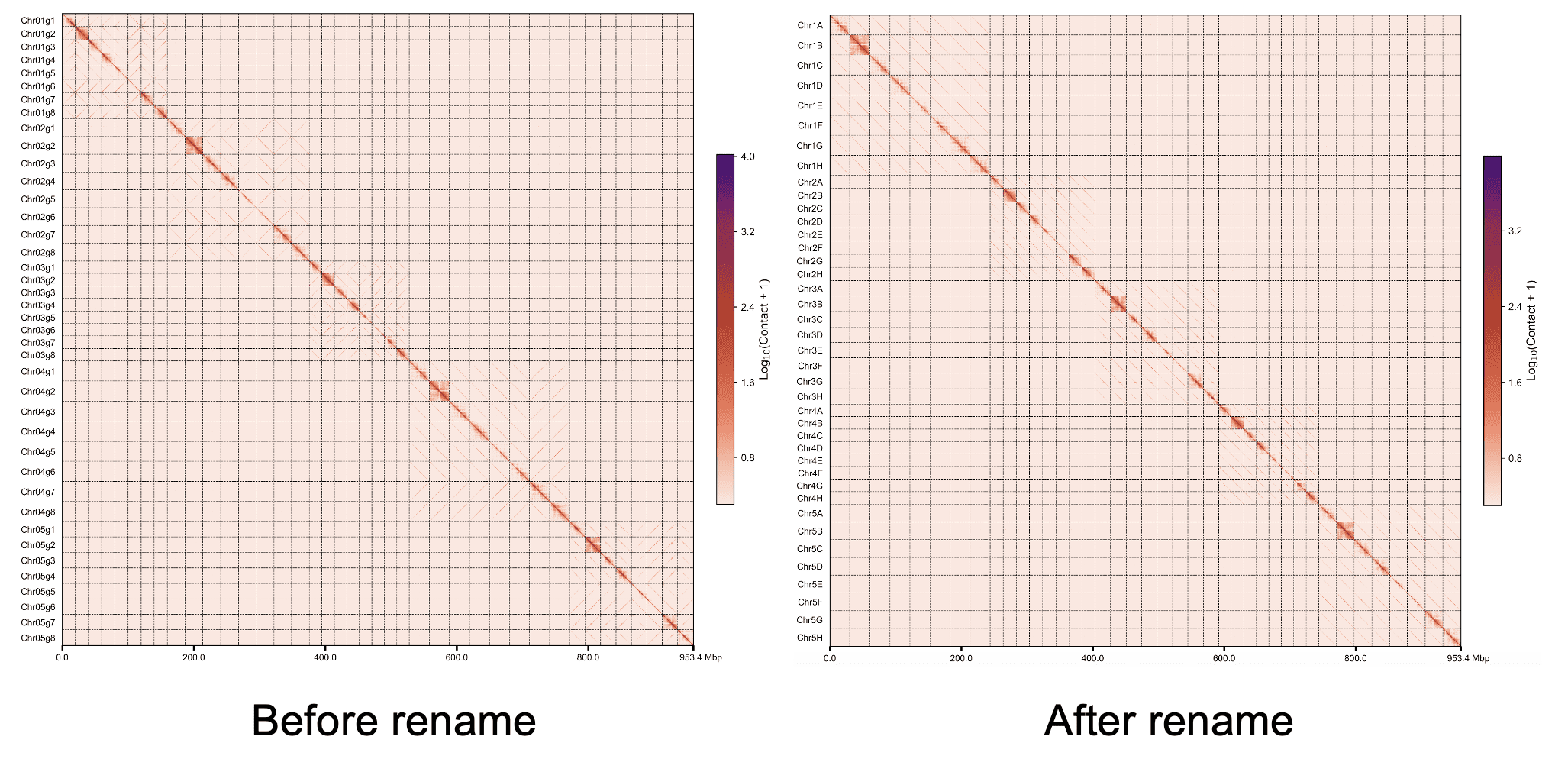
Use the uppercase letter as the suffix of the chromosome name#
Chr01A Chr01B Chr01C ...
Parameters#
Usage: cphasing rename [OPTIONS]
Rename and orientation the groups according to a refernce.
To speed up this function, we only align the first haplotype to reference, because the orientation
among different haplotypes has been paralleled in scaffolding. If you want to orient all of the
haplotypes you can specify the --unphased parameters.
╭─ Options ───────────────────────────────────────────────────────────────────────────────────────────╮
│ * --ref -r Genome of monoploid or closely related species. │
│ (REFERENCE FASTA) │
│ [required] │
│ * --fasta -f Path to draft assembly │
│ (DRAFT FASTA) │
│ [required] │
│ * --agp -a agp file. │
│ (AGP) │
│ [required] │
│ --unphased -unphased Haplotypes are not parallel aligned. Rename all of the chromosomes. │
│ --suffix-style -s The suffix style of renamed chromosome among different haplotypes. │
│ number: g1, g2, g3, ... (default) │
│ upperletter: A, B, C, ... │
│ lowerletter: a, b, c, ... │
│ (STR) │
│ [default: number] │
│ --output -o Output path of renamed agp file │
│ (OUTPUT_AGP) │
│ --threads -t Number of threads. │
│ (INT) │
│ [default: 8] │
│ --help -h,-help Show this message and exit. │
╰─────────────────────────────────────────────────────────────────────────────────────────────────────╯