Plot
示例#
从contig水平比对结果绘制热图#
- 将
pairs文件转成.cool矩阵文件
- 根据AGP文件绘制热图
根据AGP文件将contig水平的
.cool矩阵转成染色体水平,并绘制热图
Note
这步会产生两个文件一个是sample.10k.chrom.cool and sample.500k.chrom.cool,后续在agp文件不变的情况下,可以使用这两个文件进行热图的调整.
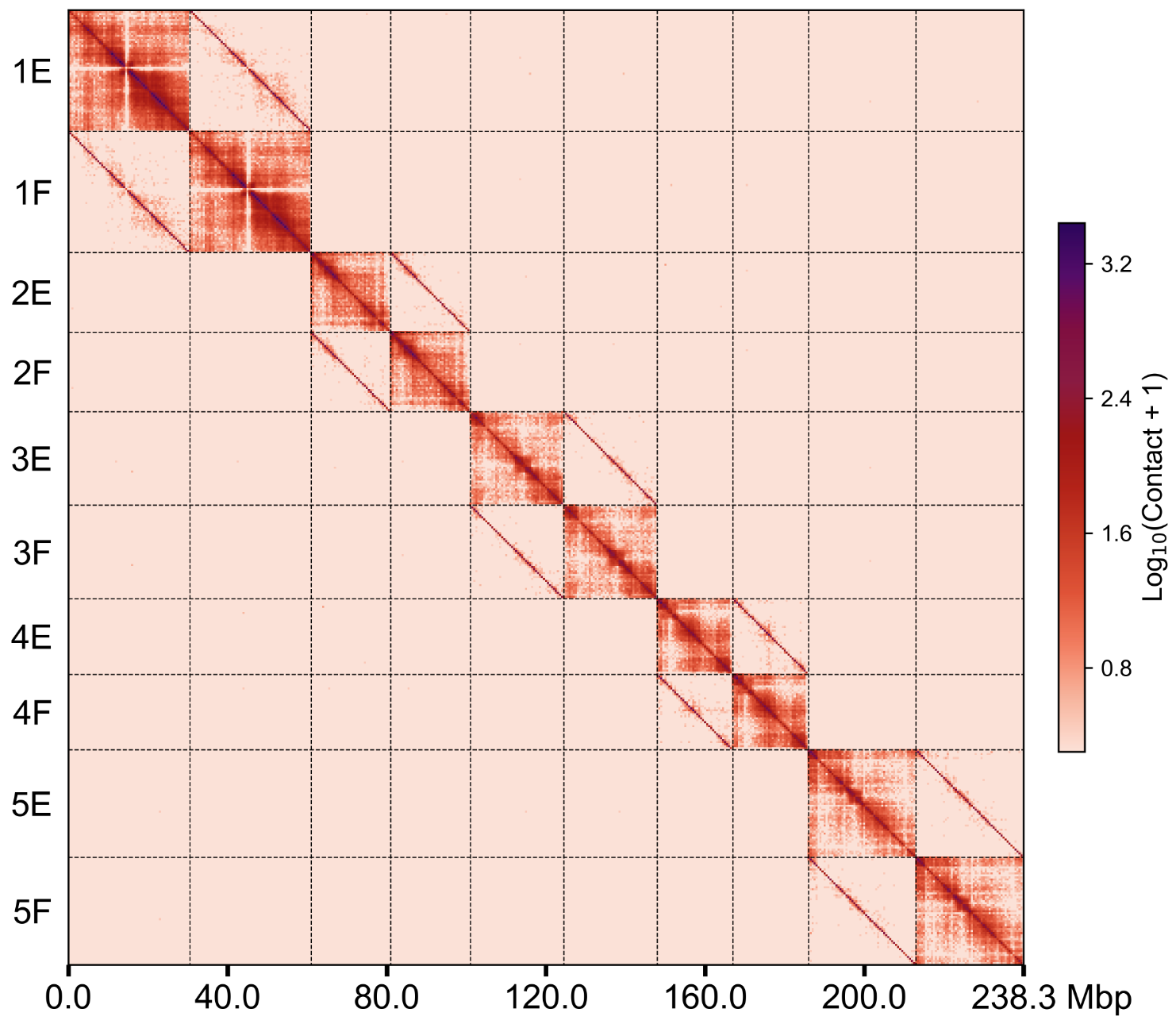
groups.500k.wg.png
直接使用.cool 文件绘制热图#
Note
用户可以指定--binsize参数来调整热图的分辨率,如果输入矩阵的分辨率跟设定的分辨率不一致,程序会自动生成新的分辨率绘制热图,但需要设定的分辨率和输入矩阵的分辨率呈整数倍的关系。例如,输入100k.cool,想绘制500k分辨率的热图:
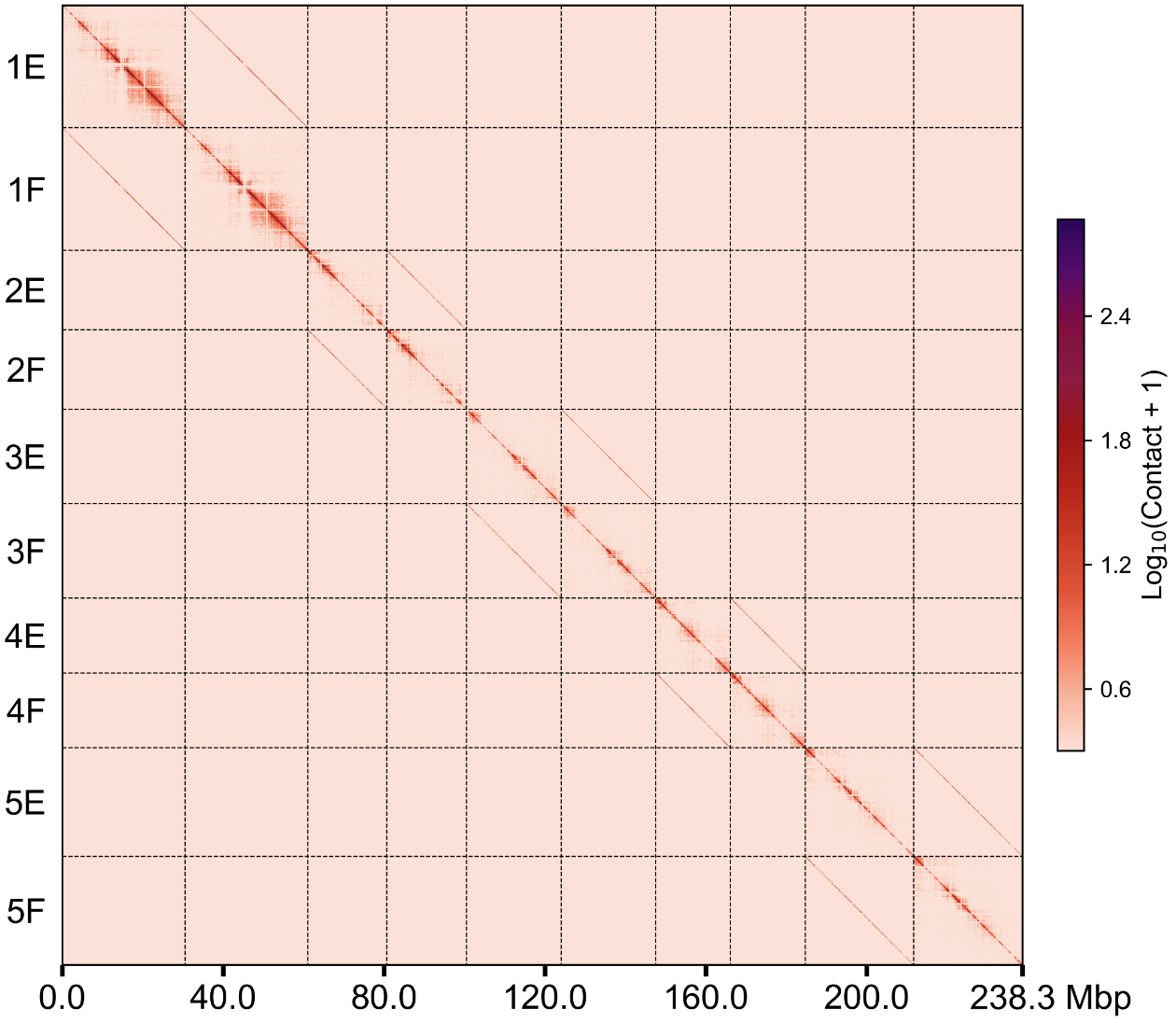

groups.100k.wg.png groups.500k.wg.png
不同参数绘制的热图#
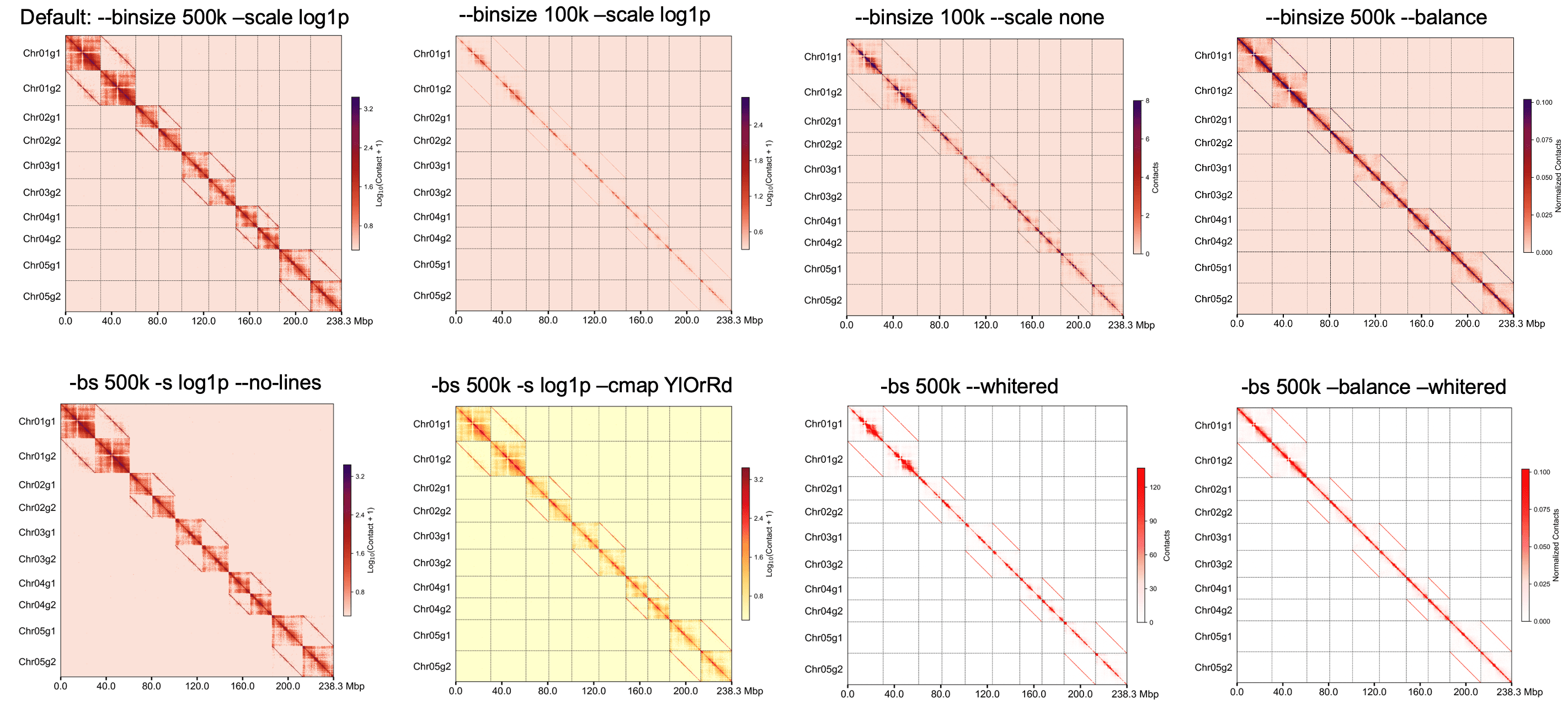
同源染色体组加边框#
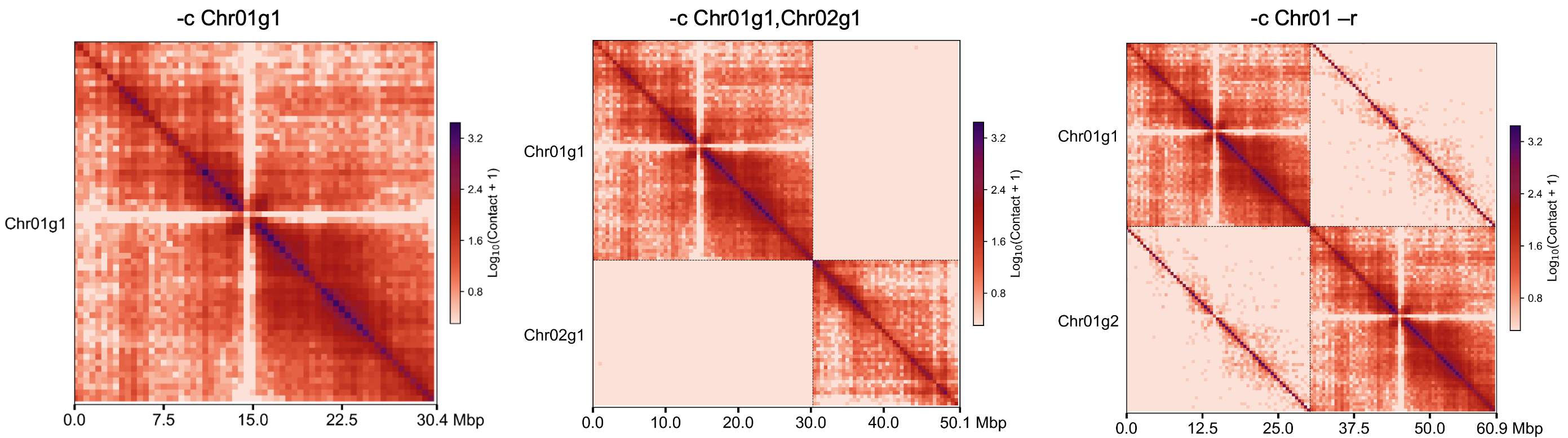
指定染色体绘制#
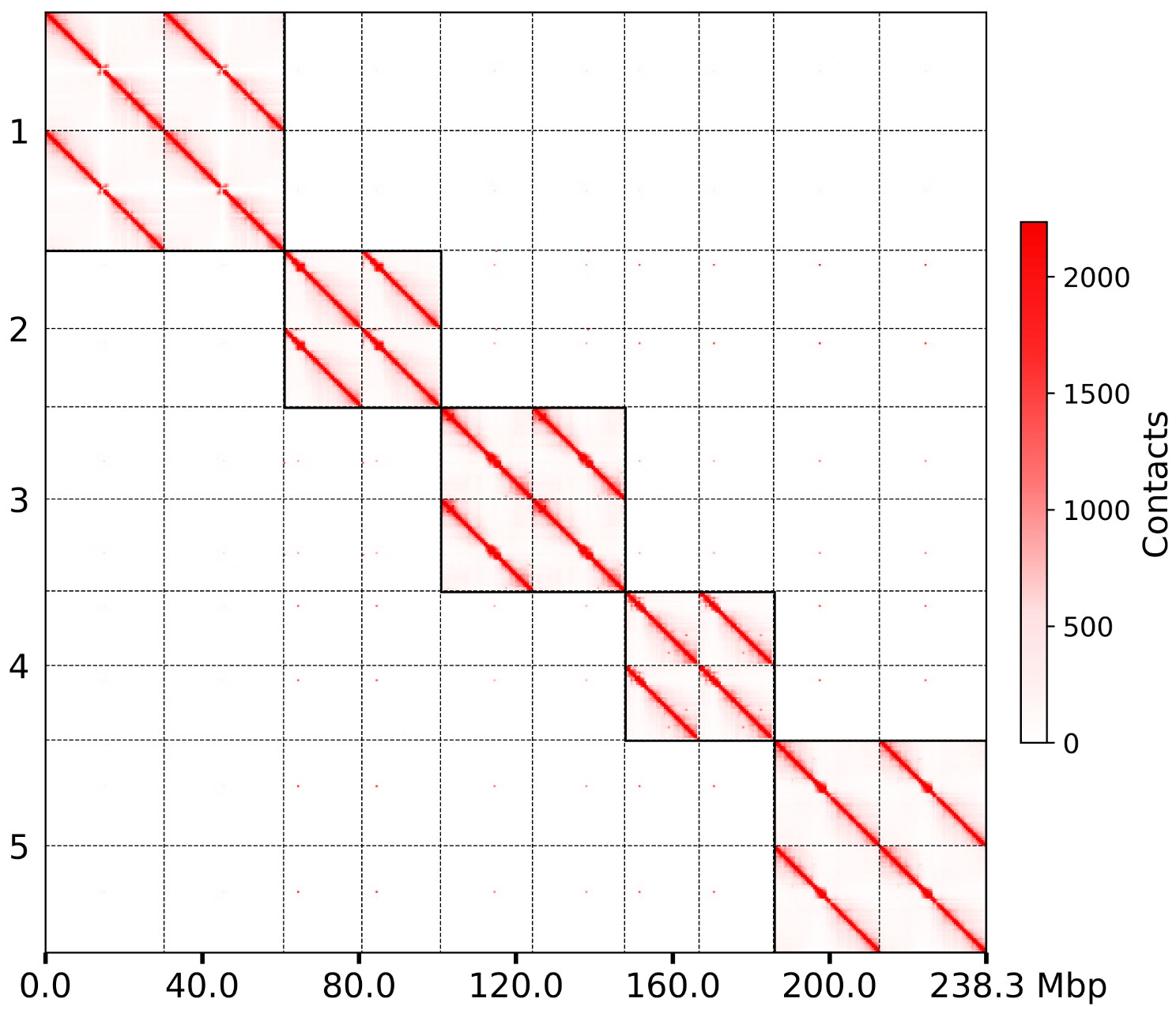
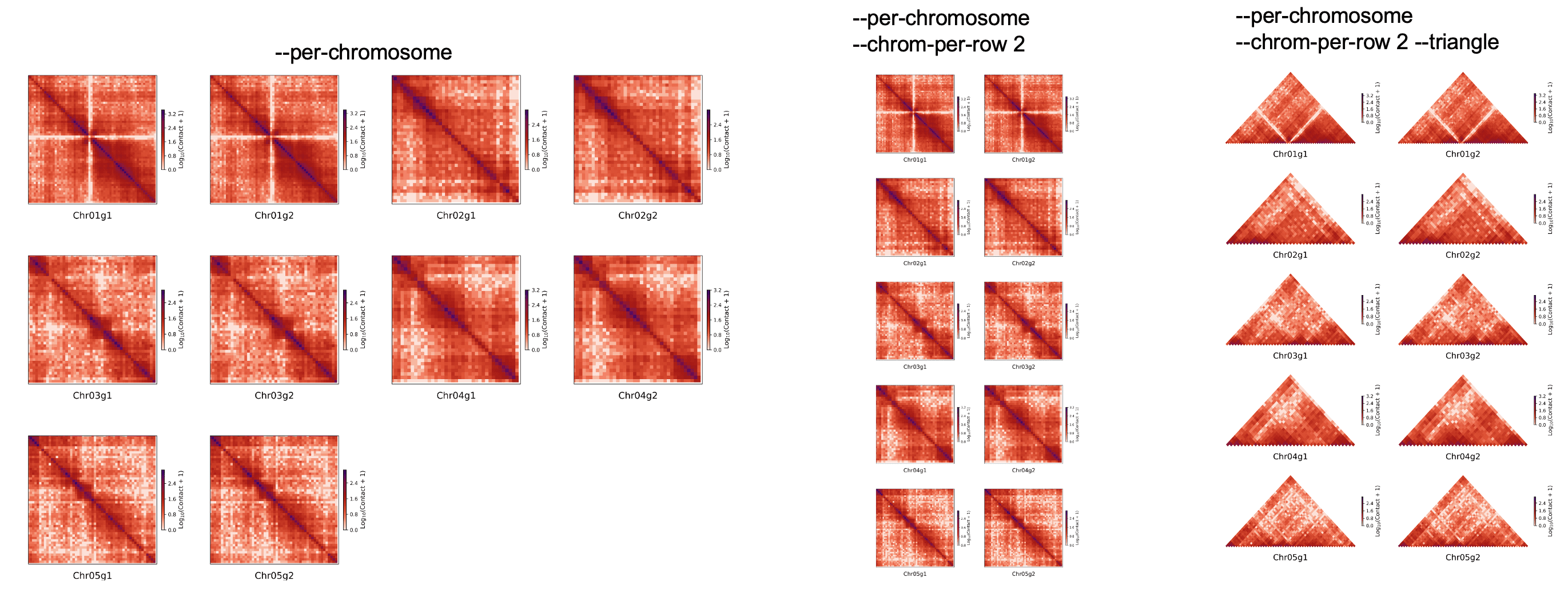
将每条染色体分开绘制#
Parameters of plot#
cphasing plot -h
Usage: cphasing plot [OPTIONS]
Adjust or Plot the contacts matrix after assembling.
▌ Usage:
▌ • adjust the matrix by agp and plot a heatmap
cphasing plot -a groups.agp -m sample.10000.cool -o groups.500k.wg.png
▌ • adjust the matrix by agp and plot a 100k resolution heatmap
cphasing plot -a groups.agp \
-m sample.10000.cool \
-o groups.100k.wg.png \
-bs 100k
▌ • only plot a heatmap
cphasing plot -m sample.100k.cool -o sample.100k.png
▌ • Plot some chromosomes
cphasing plot -m sample.100k.cool -c Chr01,Chr02 -o Chr01_Chr02.100k.png
╭─ Options of Matrix Operation ────────────────────────────────────────────╮
│ * --matrix -m Contacts matrix stored by Cool format. │
│ (COOL) │
│ [required] │
│ --binsize -bs Bin size of the heatmap you want to plot. │
│ Enabled suffix with k or m. [defalt: input │
│ matrix binsize] │
│ (STR) │
│ --only-coarsen Only coarsen the input matrix, do not need plot │
│ the heatmap. │
│ --balance balance the matrix. │
│ --balanced Plot balanced values, which need cool have │
│ weights columns in bins. │
╰──────────────────────────────────────────────────────────────────────────╯
╭─ Options of AGP Adjustment ──────────────────────────────────────────────╮
│ --agp -a (AGP) │
│ --only-adjust Only adjust the matrix by agp, do not need plot the │
│ heatmap. │
╰──────────────────────────────────────────────────────────────────────────╯
╭─ Options of Heatmap ─────────────────────────────────────────────────────╮
│ --chromosomes -c Chromosomes and order in │
│ which the chromosomes should │
│ be plotted. Comma seperated. │
│ or a one column file │
│ (TEXT) │
│ --regex -r Regular expression of │
│ chromosomes, only used for │
│ --chromosomes, e.g. Chr01g.* │
│ --disable-natural-sort -dns Disable natural sort of │
│ chromosomes, only used for │
│ --chromosomes or --only-chr │
│ --per-chromosomes,--per-chrom… -pc Instead of plotting the whole │
│ matrix, each chromosome is │
│ plotted next to the other. │
│ --only-chr -oc Only plot the chromosomes │
│ that ignore unanchored │
│ contigs. When --chromosomes │
│ specifed, this parameter will │
│ be ignored. The default use │
│ prefix of Chr to find the │
│ chromosomes. --chr-prefix can │
│ be used to change this. │
│ --chr-prefix -cp Prefix of the chromosomes, │
│ only used for --only-chr │
│ (STR) │
│ [default: Chr] │
│ --chrom-per-row -cpr Number of chromosome plot in │
│ each row │
│ (INTEGER) │
│ [default: 4] │
│ --vmin -vmin (FLOAT) │
│ --vmax -vmax (FLOAT) │
│ --scale -s Method of contact │
│ normalization │
│ (STR) │
│ [default: log1p] │
│ --triangle Plot the heatmap in triangle │
│ --fontsize Fontsize of the ticks, │
│ default is auto │
│ (INT) │
│ --dpi -dpi Resolution for the image. │
│ (INTEGER) │
│ [default: 600] │
│ --cmap,--colormap -cmap,-cm Colormap of heatmap. │
│ Available values can be seen │
│ : │
│ https://pratiman-91.github.i… │
│ and │
│ http://matplotlib.org/exampl… │
│ and whitered . │
│ (TEXT) │
│ [default: redp1_r] │
│ --whitered -wr Preset of --scale none │
│ --colormap whitered │
│ --hap-pattern -hp Regex pattern of haplotype │
│ (STR) │
│ [default: (Chr\d+)g(\d+)] │
│ --add-hap-border Add border between haplotypes,│
│ effective when --hap-pattern │
│ is set consistent with the │
│ chromosome name. │
│ --no-lines -nl Don't add dash line in │
│ chromosome boundaries. │
│ --no-ticks -nt Don't add ticks both in x │
│ axis and y axis. │
│ --rotate-xticks -rx Rotate the x ticks │
│ [default: True] │
│ --rotate-yticks -ry Rotate the x ticks │
╰──────────────────────────────────────────────────────────────────────────╯
╭─ Global Options ─────────────────────────────────────────────────────────╮
│ --output -o Output path of file. │
│ (TEXT) │
│ [default: plot.heatmap.png] │
│ --threads -t Number of threads. Used in matrix balance. │
│ (INT) │
│ [default: 8] │
│ --help -help,-h Show this message and exit. │
╰──────────────────────────────────────────────────────────────────────────╯