Plot
Examples¶
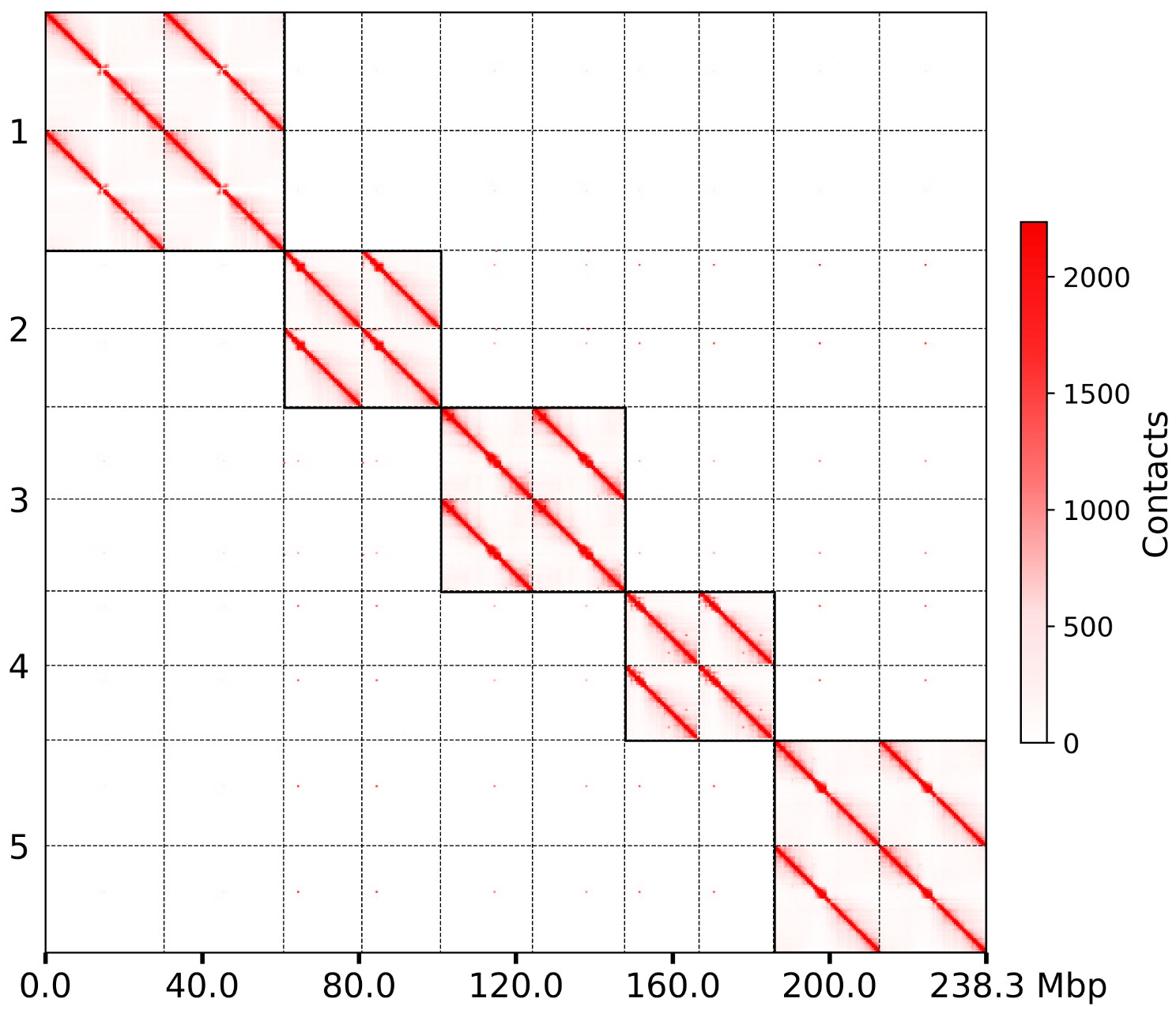
Plot chromosome-level heatmap from contig-level mapping result¶
- Convert pairs to cool file
- Adjust contig-level matrix to chromosome-level by agp file
Note
This function will generate two intermedia sample.10000.chrom.cool and sample.500k.chrom.cool.
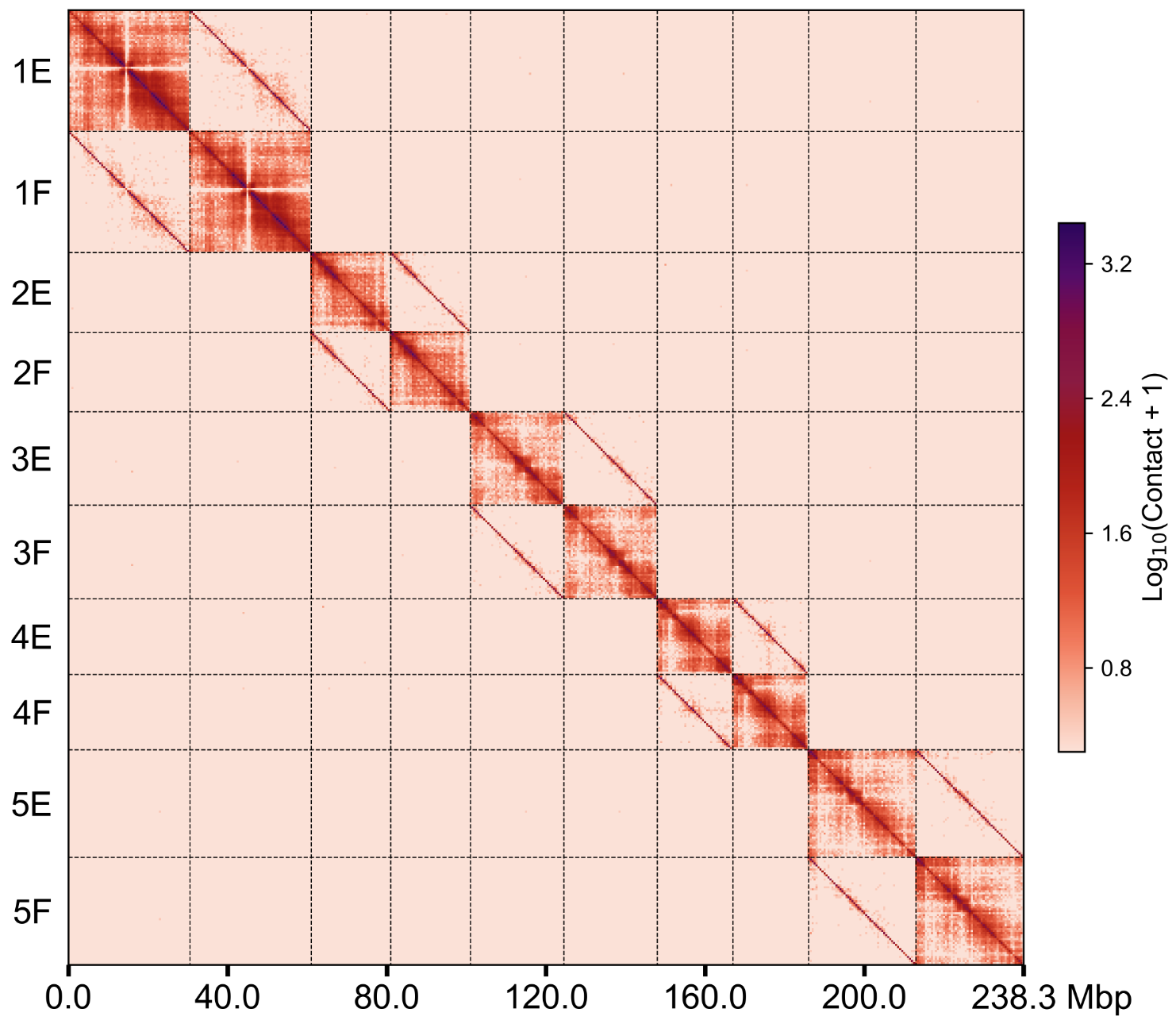
groups.500k.wg.png
Directly plot heatmap from .cool file¶
Note
--binsize can be specified when you want to plot a larger resolution, e.g., 500k, plot will automatically generate a 1m matrix from the input matrix. Still, the binsize of the output matrix should be in integer multiples than the binsize of the input matrix.
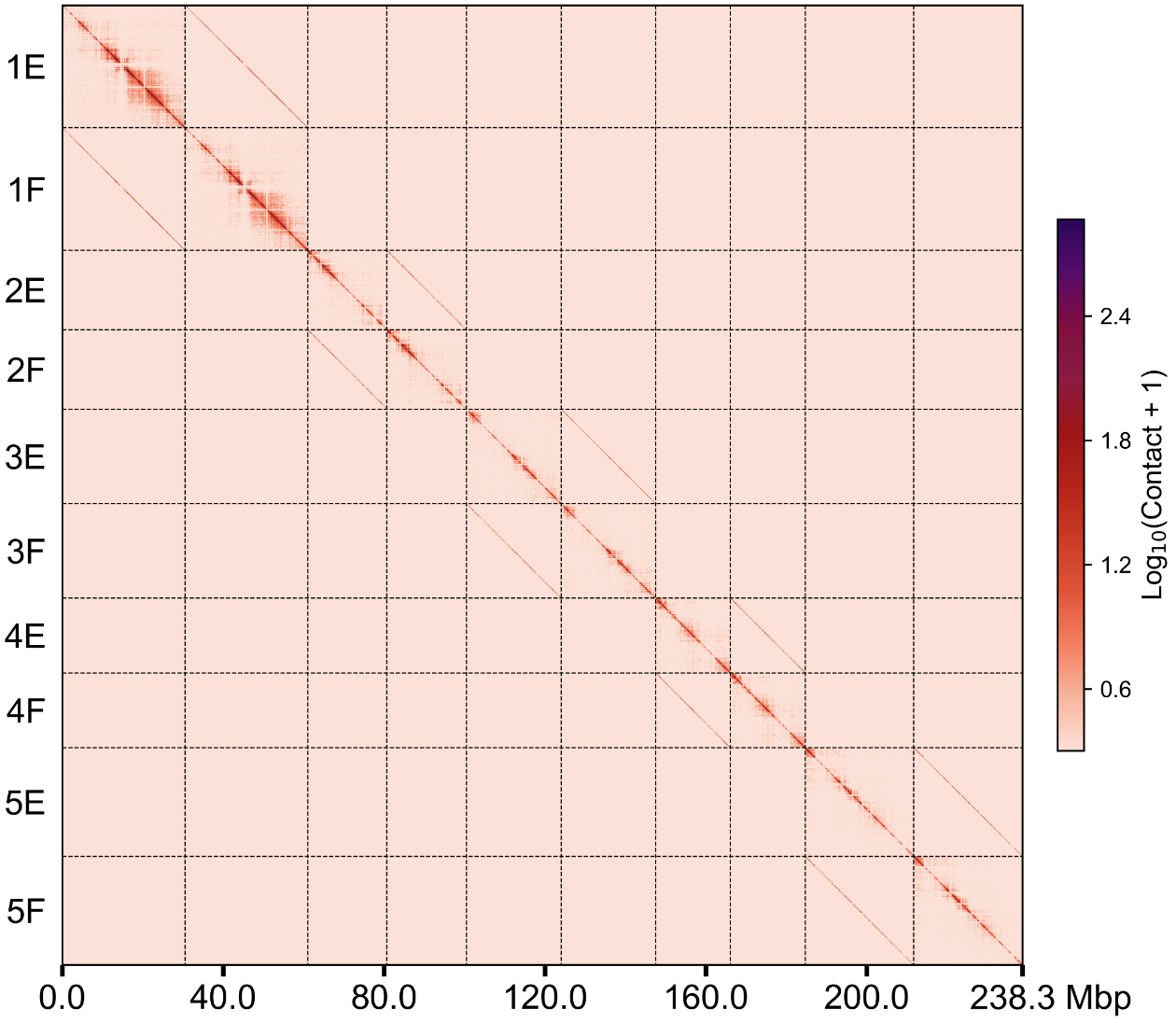

groups.100k.wg.png groups.500k.wg.png
Gallery of different parameters¶
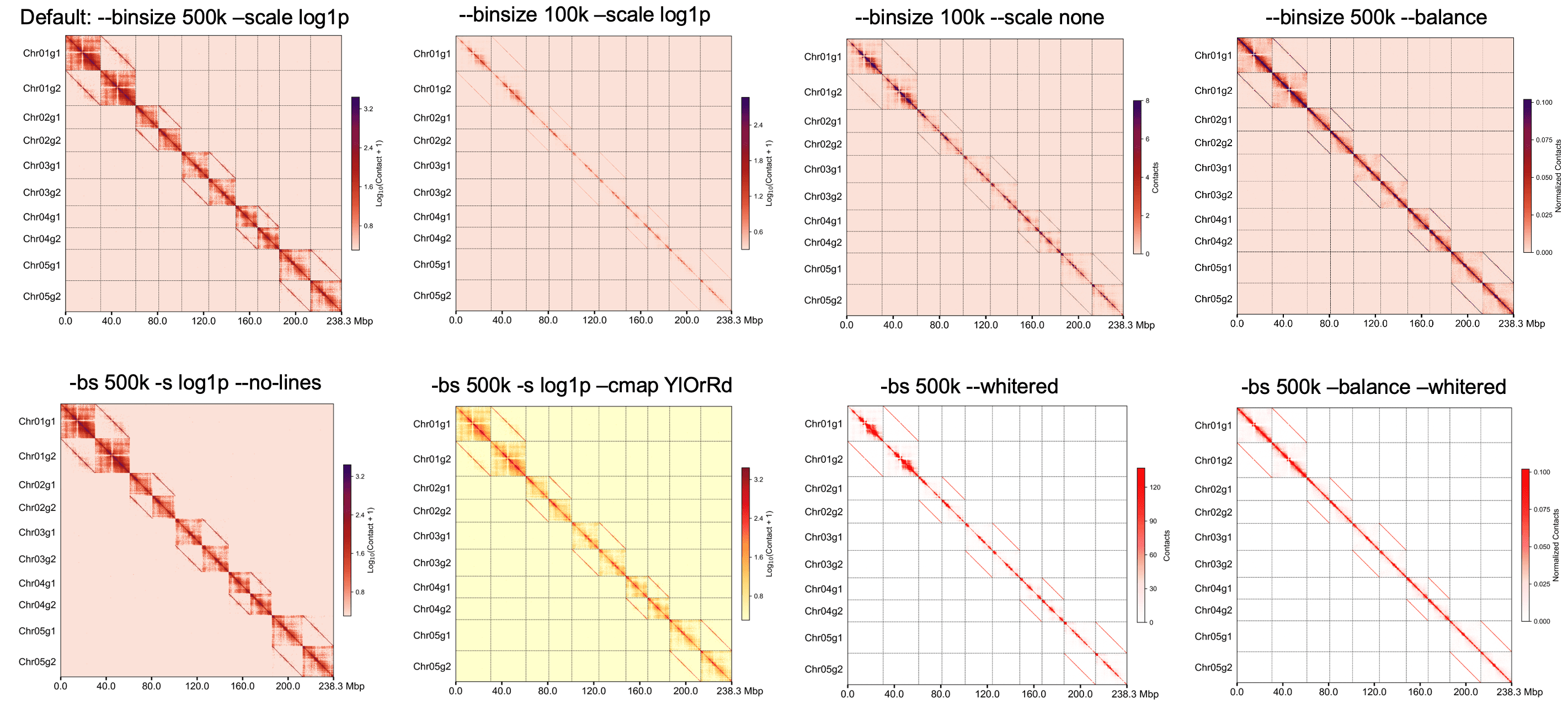
Add line to highlight the haplotype border¶
Plot specified chromosome¶
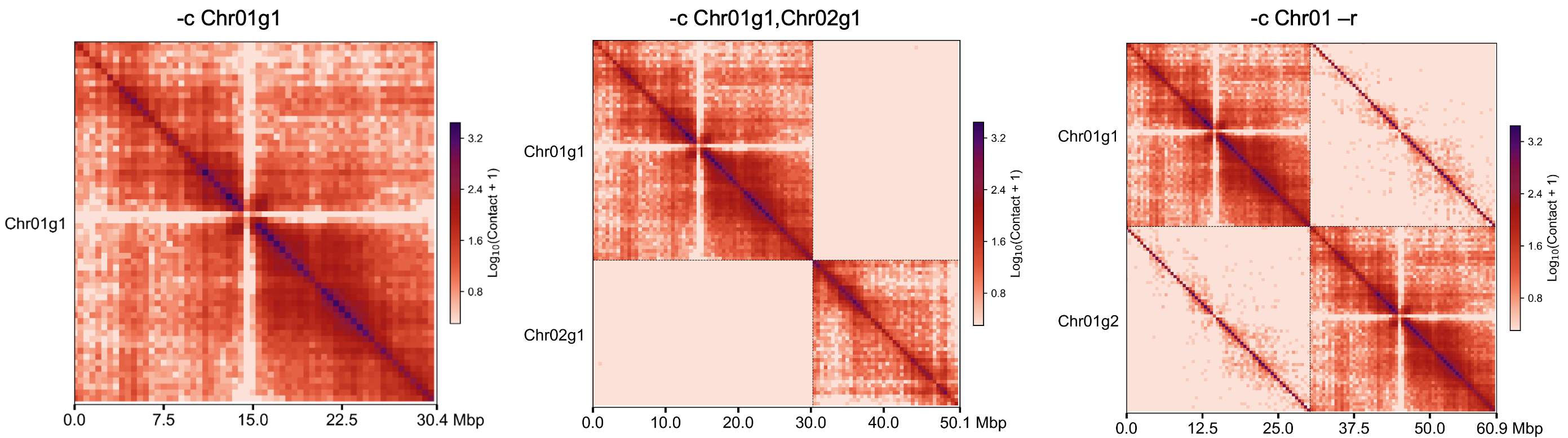
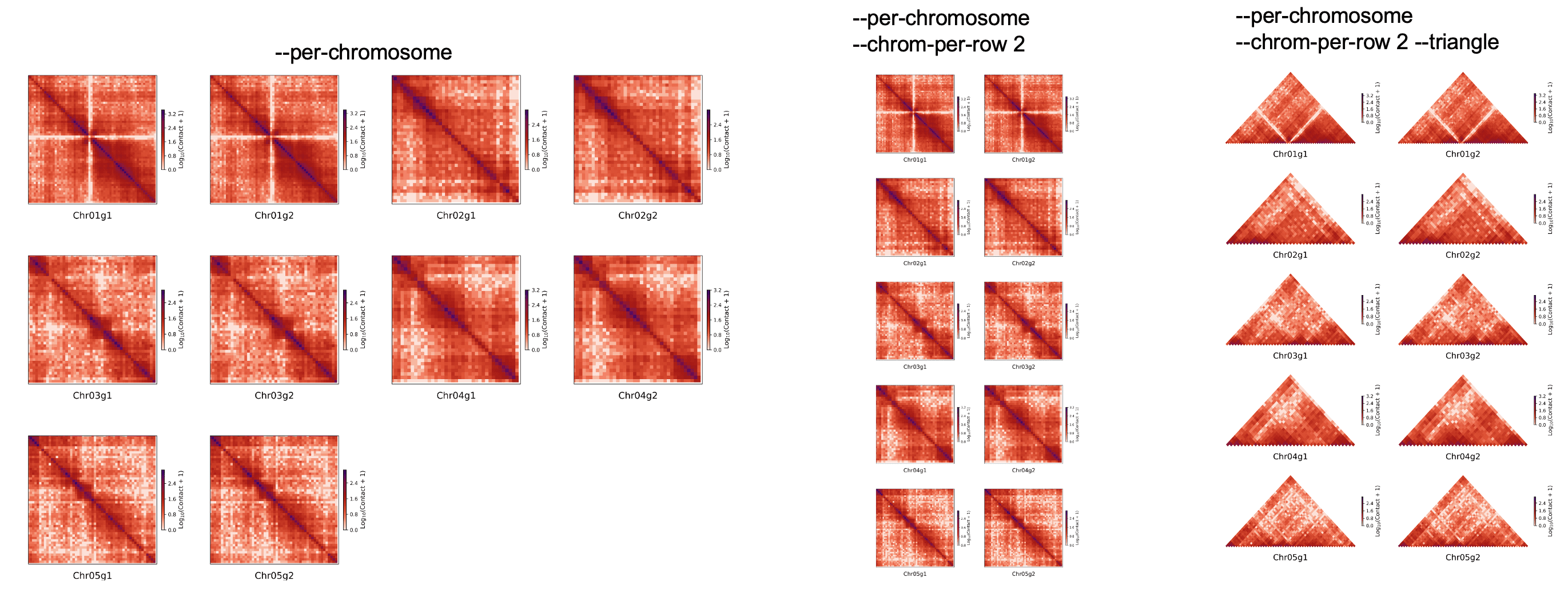
Plot all chromosome split in one picture¶