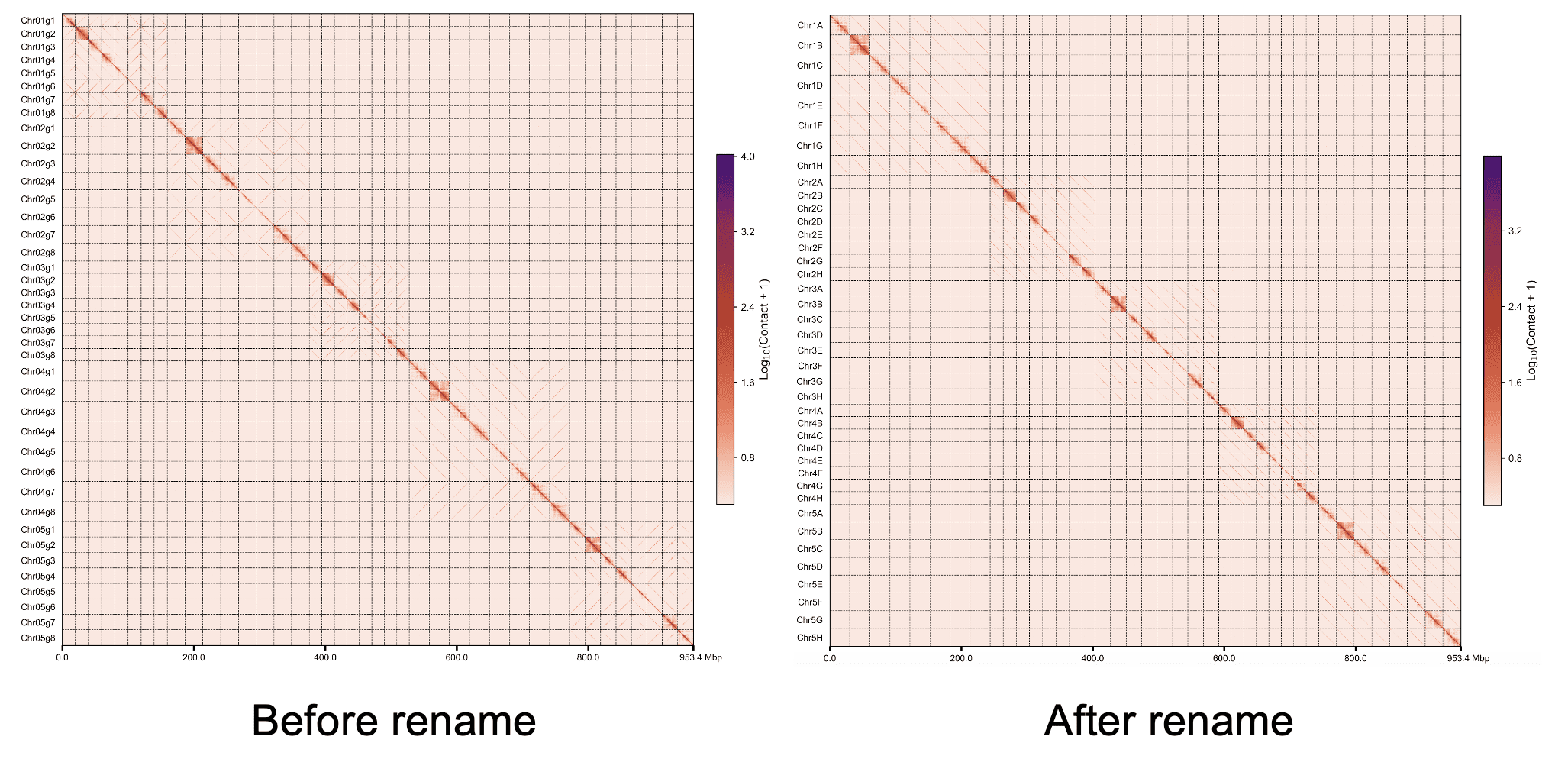
Rename¶
以单倍型或者近缘物种的参考基因组作为参考,将当前组装的多倍体进行重命名和染色体方向调整.
调整phasing mode的组装结果¶
默认情况下phasing mode已经将同源染色体聚成一组,并且不同同源染色体将的方向已经调整至平行,为了节省时间开销,我们仅将第一个单倍型即Chr??.g1比对至参考基因组上,剩下的单倍型命名和方向跟随g1进行调整。如果不符合以上说明,则需加--unphased参数对所有染色体进行比对。
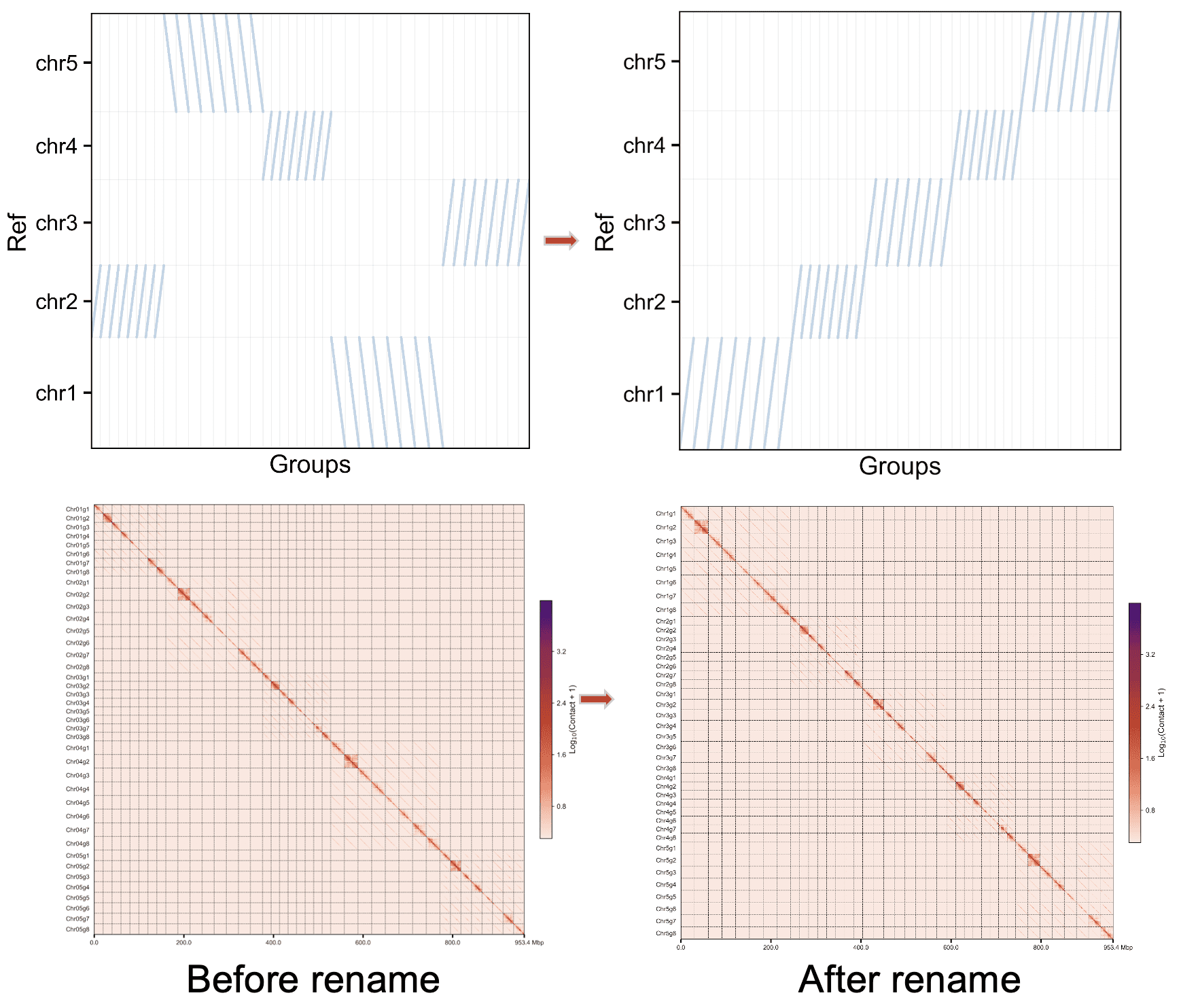
对所有染色体进行重命名¶
basal或者basal_withprune mode,需要对所有染色体进行比对,然后重命名和重新调整方向,需要加--unphased参数。
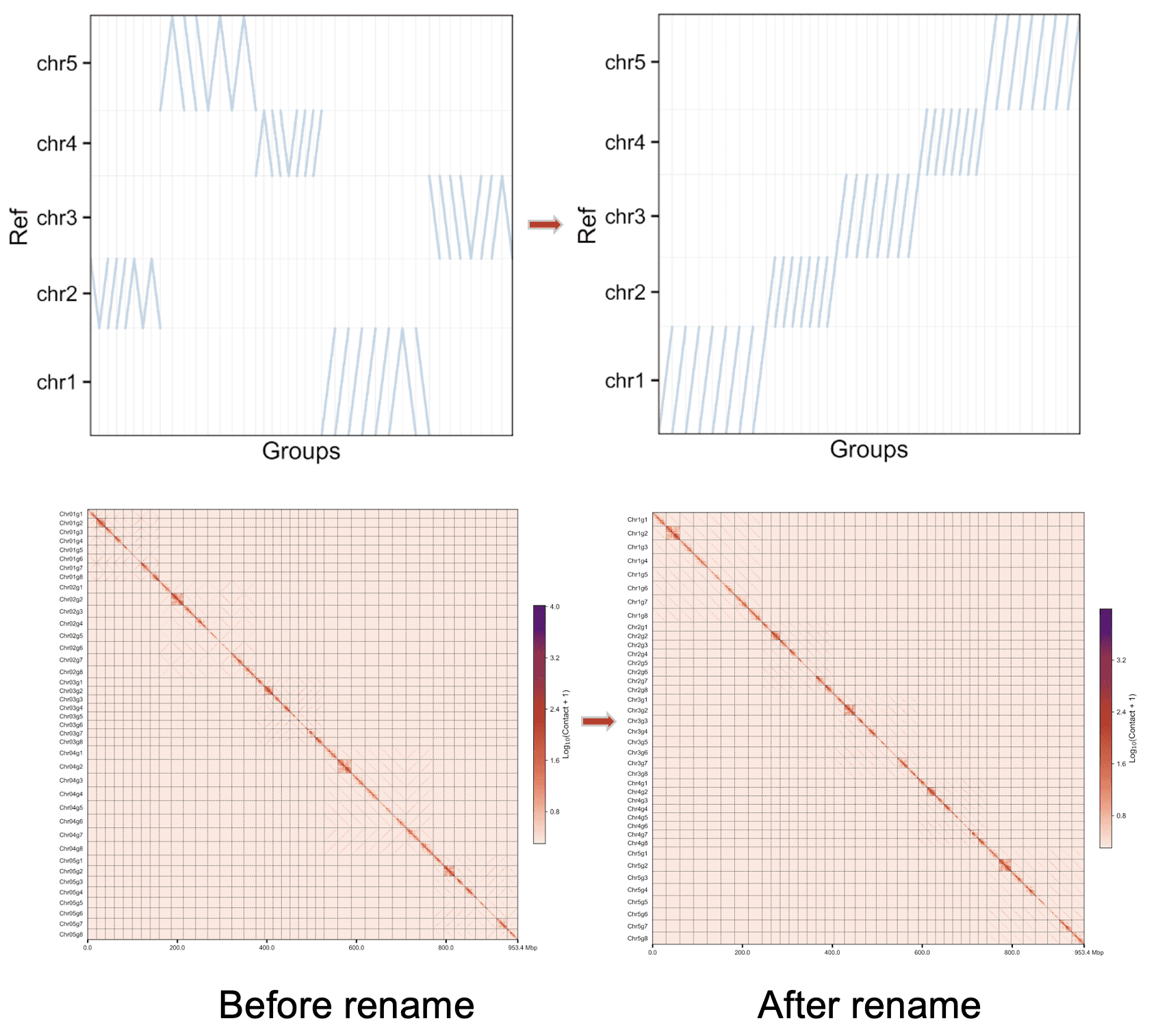
用字母作为不同单倍型的后缀¶
Chr01A Chr01B Chr01C ...